Chapter 10 Working with multiple datasets
R
library(tidyverse)
library(janitor)
encounters <- read_csv("data/synthea/encounters.csv") %>%
clean_names()encounters_mi <- encounters %>%
filter(str_to_lower(description) %in%
str_to_lower(c("Cardiac Arrest", "Myocardial Infarction"))
)
encounters_mi# A tibble: 70 × 15
id start stop patient organization provider
<chr> <dttm> <dttm> <chr> <chr> <chr>
1 2500b… 2001-07-04 08:42:44 2001-07-04 10:27:44 d49f748… 23834663-ed… af42f30…
2 83cda… 2010-10-24 05:28:51 2010-10-24 07:13:51 e05dd03… 465de31f-30… 0a8a935…
3 7302a… 2017-06-04 11:11:47 2017-06-04 12:56:47 0f5646b… 4861d01f-01… b08f34d…
4 7db01… 1989-01-17 18:12:03 1989-01-17 19:57:03 44c8b4c… b0e04623-b0… 58b66cc…
5 e73fe… 1973-07-19 04:25:46 1973-07-19 06:10:46 8975205… ef6ab57c-ed… 77a7881…
6 4f126… 2017-07-13 04:42:48 2017-07-13 06:27:48 8d1ba4b… ef58ea08-d8… 3421aa7…
7 00d8f… 2007-08-23 19:03:06 2007-08-23 20:48:06 87be3f6… 08bcda9c-f8… 01c46a2…
8 1785e… 2010-03-28 05:41:47 2010-03-28 07:26:47 fcd3f56… 12c9daf5-a2… aa89beb…
9 f026c… 1991-02-18 01:51:56 1991-02-18 03:36:56 47392cc… 5d4b9df1-93… af01a38…
10 20083… 2017-03-21 09:16:22 2017-03-21 11:01:22 0447625… e44f438a-60… de75d49…
# … with 60 more rows, and 9 more variables: payer <chr>, encounterclass <chr>,
# code <dbl>, description <chr>, base_encounter_cost <dbl>,
# total_claim_cost <dbl>, payer_coverage <dbl>, reasoncode <dbl>,
# reasondescription <chr>Q: How old was the patient at the time of a “heart attack”?
R
patients <- read_csv("data/synthea/patients.csv") %>%
clean_names()Rows: 1171 Columns: 25── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (19): Id, SSN, DRIVERS, PASSPORT, PREFIX, FIRST, LAST, SUFFIX, MAIDEN, ...
dbl (4): LAT, LON, HEALTHCARE_EXPENSES, HEALTHCARE_COVERAGE
date (2): BIRTHDATE, DEATHDATE
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.patients# A tibble: 1,171 × 25
id birthdate deathdate ssn drivers passport prefix first last suffix
<chr> <date> <date> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 1d604d… 1989-05-25 NA 999-… S99984… X192772… Mr. José… Góme… <NA>
2 034e9e… 1983-11-14 NA 999-… S99962… X882754… Mr. Milo… Feil… <NA>
3 10339b… 1992-06-02 NA 999-… S99972… X737544… Mr. Jays… Fade… <NA>
4 8d4c43… 1978-05-27 NA 999-… S99974… X409155… Mrs. Mari… Ruth… <NA>
5 f5dcd4… 1996-10-18 NA 999-… S99915… X867729… Mr. Greg… Auer… <NA>
6 72c0b9… 2017-07-27 NA 999-… <NA> <NA> <NA> Jaci… Kris… <NA>
7 b1e9b0… 2003-12-13 NA 999-… S99954… <NA> <NA> Jimm… Harr… <NA>
8 01207e… 2019-05-15 NA 999-… <NA> <NA> <NA> Kary… Muel… <NA>
9 b58731… 1970-05-16 NA 999-… S99978… X781703… Mrs. Isab… Luci… <NA>
10 cfee79… 2016-07-04 NA 999-… <NA> <NA> <NA> Alva… Kraj… <NA>
# … with 1,161 more rows, and 15 more variables: maiden <chr>, marital <chr>,
# race <chr>, ethnicity <chr>, gender <chr>, birthplace <chr>, address <chr>,
# city <chr>, state <chr>, county <chr>, zip <chr>, lat <dbl>, lon <dbl>,
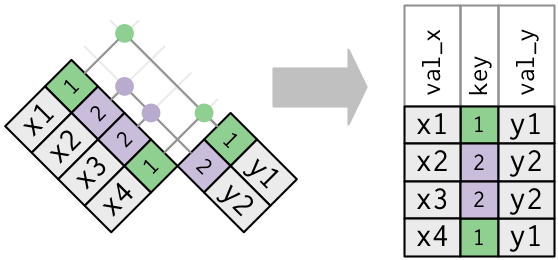
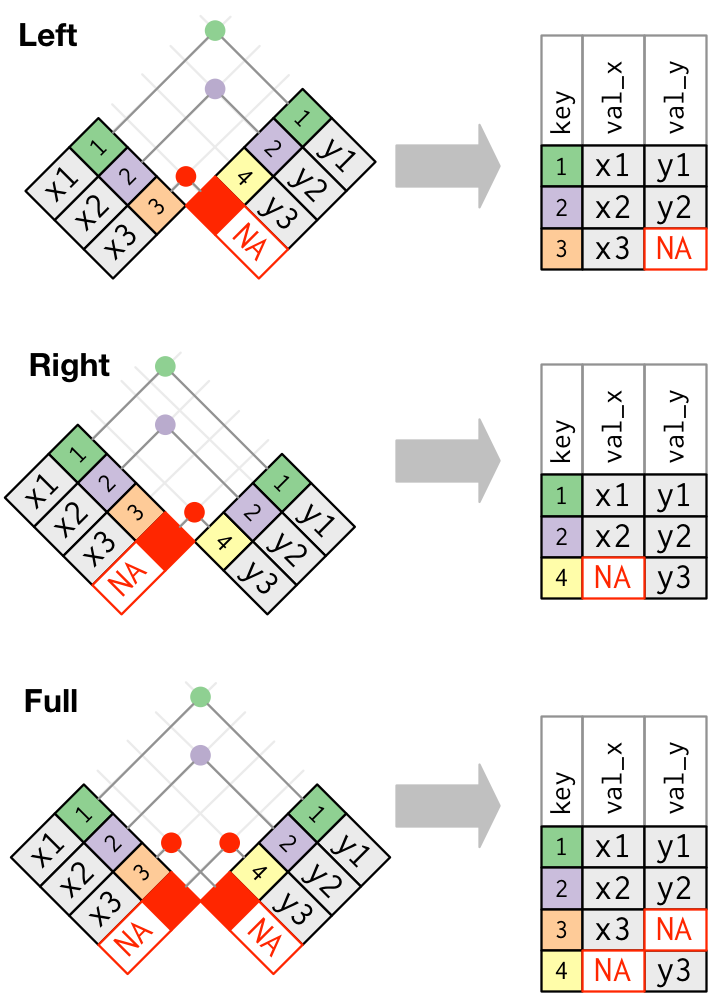
# healthcare_expenses <dbl>, healthcare_coverage <dbl>10.1 Joins
https://r4ds.had.co.nz/relational-data.html
inner_join(): includes all rows in x and y.
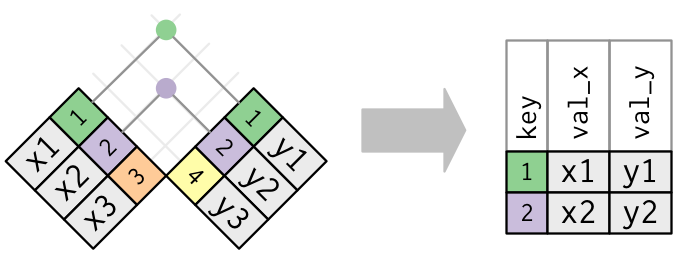
left_join(): includes all rows in x.right_join(): includes all rows in y.full_join(): includes all rows in x or y.
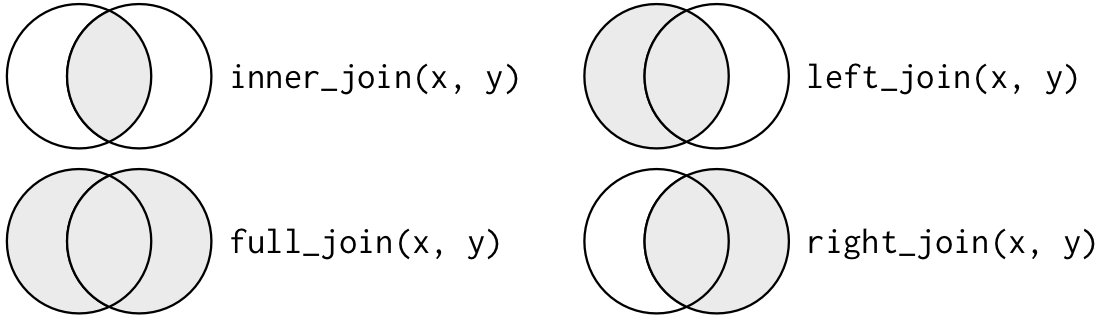
encounters_pt <- encounters_mi %>%
left_join(patients, by = c("patient" = "id"))
encounters_pt# A tibble: 70 × 39
id start stop patient organization provider
<chr> <dttm> <dttm> <chr> <chr> <chr>
1 2500b… 2001-07-04 08:42:44 2001-07-04 10:27:44 d49f748… 23834663-ed… af42f30…
2 83cda… 2010-10-24 05:28:51 2010-10-24 07:13:51 e05dd03… 465de31f-30… 0a8a935…
3 7302a… 2017-06-04 11:11:47 2017-06-04 12:56:47 0f5646b… 4861d01f-01… b08f34d…
4 7db01… 1989-01-17 18:12:03 1989-01-17 19:57:03 44c8b4c… b0e04623-b0… 58b66cc…
5 e73fe… 1973-07-19 04:25:46 1973-07-19 06:10:46 8975205… ef6ab57c-ed… 77a7881…
6 4f126… 2017-07-13 04:42:48 2017-07-13 06:27:48 8d1ba4b… ef58ea08-d8… 3421aa7…
7 00d8f… 2007-08-23 19:03:06 2007-08-23 20:48:06 87be3f6… 08bcda9c-f8… 01c46a2…
8 1785e… 2010-03-28 05:41:47 2010-03-28 07:26:47 fcd3f56… 12c9daf5-a2… aa89beb…
9 f026c… 1991-02-18 01:51:56 1991-02-18 03:36:56 47392cc… 5d4b9df1-93… af01a38…
10 20083… 2017-03-21 09:16:22 2017-03-21 11:01:22 0447625… e44f438a-60… de75d49…
# … with 60 more rows, and 33 more variables: payer <chr>,
# encounterclass <chr>, code <dbl>, description <chr>,
# base_encounter_cost <dbl>, total_claim_cost <dbl>, payer_coverage <dbl>,
# reasoncode <dbl>, reasondescription <chr>, birthdate <date>,
# deathdate <date>, ssn <chr>, drivers <chr>, passport <chr>, prefix <chr>,
# first <chr>, last <chr>, suffix <chr>, maiden <chr>, marital <chr>,
# race <chr>, ethnicity <chr>, gender <chr>, birthplace <chr>, …10.1.2 Formally test your assumptions
R
stopifnot(nrow(encounters_mi) == nrow(encounters_pt)) # test your workstopifnot(nrow(encounters_mi) == 100) # test your workError: nrow(encounters_mi) == 100 is not TRUE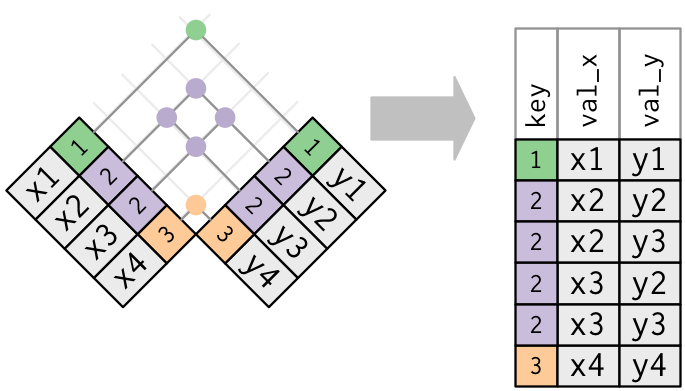
10.1.3 Different types of joins
R
pt_encounters <- patients %>%
left_join(encounters_mi, by = c("id" = "patient"))
pt_encounters# A tibble: 1,175 × 39
id birthdate deathdate ssn drivers passport prefix first last suffix
<chr> <date> <date> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 1d604d… 1989-05-25 NA 999-… S99984… X192772… Mr. José… Góme… <NA>
2 034e9e… 1983-11-14 NA 999-… S99962… X882754… Mr. Milo… Feil… <NA>
3 10339b… 1992-06-02 NA 999-… S99972… X737544… Mr. Jays… Fade… <NA>
4 8d4c43… 1978-05-27 NA 999-… S99974… X409155… Mrs. Mari… Ruth… <NA>
5 f5dcd4… 1996-10-18 NA 999-… S99915… X867729… Mr. Greg… Auer… <NA>
6 72c0b9… 2017-07-27 NA 999-… <NA> <NA> <NA> Jaci… Kris… <NA>
7 b1e9b0… 2003-12-13 NA 999-… S99954… <NA> <NA> Jimm… Harr… <NA>
8 01207e… 2019-05-15 NA 999-… <NA> <NA> <NA> Kary… Muel… <NA>
9 b58731… 1970-05-16 NA 999-… S99978… X781703… Mrs. Isab… Luci… <NA>
10 cfee79… 2016-07-04 NA 999-… <NA> <NA> <NA> Alva… Kraj… <NA>
# … with 1,165 more rows, and 29 more variables: maiden <chr>, marital <chr>,
# race <chr>, ethnicity <chr>, gender <chr>, birthplace <chr>, address <chr>,
# city <chr>, state <chr>, county <chr>, zip <chr>, lat <dbl>, lon <dbl>,
# healthcare_expenses <dbl>, healthcare_coverage <dbl>, id.y <chr>,
# start <dttm>, stop <dttm>, organization <chr>, provider <chr>, payer <chr>,
# encounterclass <chr>, code <dbl>, description <chr>,
# base_encounter_cost <dbl>, total_claim_cost <dbl>, payer_coverage <dbl>, …inner_join(encounters_mi, patients, by = c("patient" = "id"))# A tibble: 70 × 39
id start stop patient organization provider
<chr> <dttm> <dttm> <chr> <chr> <chr>
1 2500b… 2001-07-04 08:42:44 2001-07-04 10:27:44 d49f748… 23834663-ed… af42f30…
2 83cda… 2010-10-24 05:28:51 2010-10-24 07:13:51 e05dd03… 465de31f-30… 0a8a935…
3 7302a… 2017-06-04 11:11:47 2017-06-04 12:56:47 0f5646b… 4861d01f-01… b08f34d…
4 7db01… 1989-01-17 18:12:03 1989-01-17 19:57:03 44c8b4c… b0e04623-b0… 58b66cc…
5 e73fe… 1973-07-19 04:25:46 1973-07-19 06:10:46 8975205… ef6ab57c-ed… 77a7881…
6 4f126… 2017-07-13 04:42:48 2017-07-13 06:27:48 8d1ba4b… ef58ea08-d8… 3421aa7…
7 00d8f… 2007-08-23 19:03:06 2007-08-23 20:48:06 87be3f6… 08bcda9c-f8… 01c46a2…
8 1785e… 2010-03-28 05:41:47 2010-03-28 07:26:47 fcd3f56… 12c9daf5-a2… aa89beb…
9 f026c… 1991-02-18 01:51:56 1991-02-18 03:36:56 47392cc… 5d4b9df1-93… af01a38…
10 20083… 2017-03-21 09:16:22 2017-03-21 11:01:22 0447625… e44f438a-60… de75d49…
# … with 60 more rows, and 33 more variables: payer <chr>,
# encounterclass <chr>, code <dbl>, description <chr>,
# base_encounter_cost <dbl>, total_claim_cost <dbl>, payer_coverage <dbl>,
# reasoncode <dbl>, reasondescription <chr>, birthdate <date>,
# deathdate <date>, ssn <chr>, drivers <chr>, passport <chr>, prefix <chr>,
# first <chr>, last <chr>, suffix <chr>, maiden <chr>, marital <chr>,
# race <chr>, ethnicity <chr>, gender <chr>, birthplace <chr>, …full_join(encounters_mi, patients, by = c("patient" = "id"))# A tibble: 1,175 × 39
id start stop patient organization provider
<chr> <dttm> <dttm> <chr> <chr> <chr>
1 2500b… 2001-07-04 08:42:44 2001-07-04 10:27:44 d49f748… 23834663-ed… af42f30…
2 83cda… 2010-10-24 05:28:51 2010-10-24 07:13:51 e05dd03… 465de31f-30… 0a8a935…
3 7302a… 2017-06-04 11:11:47 2017-06-04 12:56:47 0f5646b… 4861d01f-01… b08f34d…
4 7db01… 1989-01-17 18:12:03 1989-01-17 19:57:03 44c8b4c… b0e04623-b0… 58b66cc…
5 e73fe… 1973-07-19 04:25:46 1973-07-19 06:10:46 8975205… ef6ab57c-ed… 77a7881…
6 4f126… 2017-07-13 04:42:48 2017-07-13 06:27:48 8d1ba4b… ef58ea08-d8… 3421aa7…
7 00d8f… 2007-08-23 19:03:06 2007-08-23 20:48:06 87be3f6… 08bcda9c-f8… 01c46a2…
8 1785e… 2010-03-28 05:41:47 2010-03-28 07:26:47 fcd3f56… 12c9daf5-a2… aa89beb…
9 f026c… 1991-02-18 01:51:56 1991-02-18 03:36:56 47392cc… 5d4b9df1-93… af01a38…
10 20083… 2017-03-21 09:16:22 2017-03-21 11:01:22 0447625… e44f438a-60… de75d49…
# … with 1,165 more rows, and 33 more variables: payer <chr>,
# encounterclass <chr>, code <dbl>, description <chr>,
# base_encounter_cost <dbl>, total_claim_cost <dbl>, payer_coverage <dbl>,
# reasoncode <dbl>, reasondescription <chr>, birthdate <date>,
# deathdate <date>, ssn <chr>, drivers <chr>, passport <chr>, prefix <chr>,
# first <chr>, last <chr>, suffix <chr>, maiden <chr>, marital <chr>,
# race <chr>, ethnicity <chr>, gender <chr>, birthplace <chr>, …10.1.4 Age at time of heart attack
R
library(lubridate)
Attaching package: 'lubridate'The following objects are masked from 'package:base':
date, intersect, setdiff, uniontoday()[1] "2021-10-09"current_date = ymd("2020-04-28")
current_date[1] "2020-04-28"10.1.4.1 Date arithmetic
R
# age "today"
mi_encounters_pt <- encounters_pt %>%
select(id, start, stop, patient, description, reasondescription, birthdate, deathdate) %>%
mutate(age_today_duration = birthdate %--% current_date,
age_years_today = trunc(as.duration(age_today_duration) / dyears(1))
)# age at event
mi_encounters_pt <- mi_encounters_pt %>%
mutate(age_event_duration = birthdate %--% start,
age_years_event = trunc(as.duration(age_event_duration) / dyears(1))
)R
library(ggplot2)
ggplot(mi_encounters_pt, aes(x = age_years_event)) +
geom_histogram(bins = 10)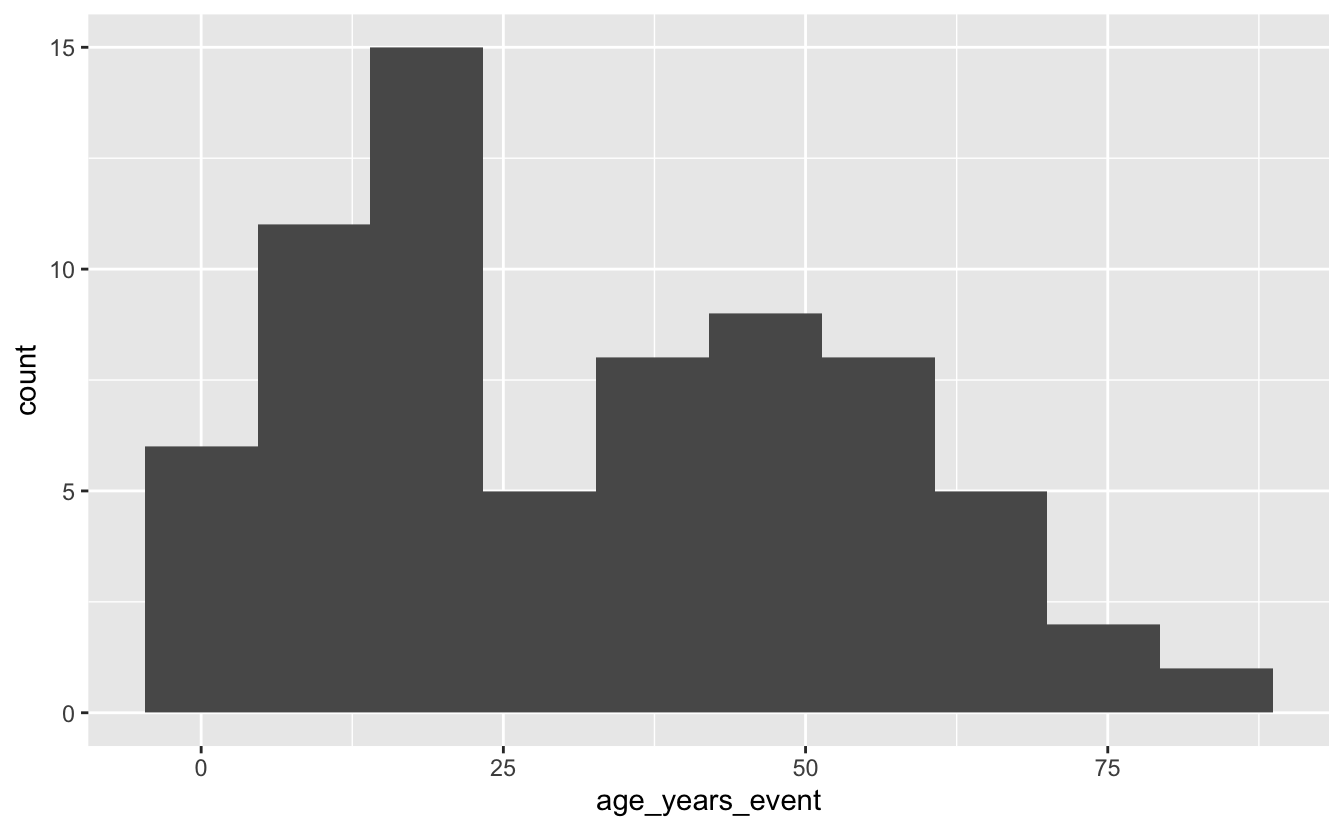
ggplot(mi_encounters_pt, aes(x = age_years_event, fill=description)) +
geom_histogram(bins = 10)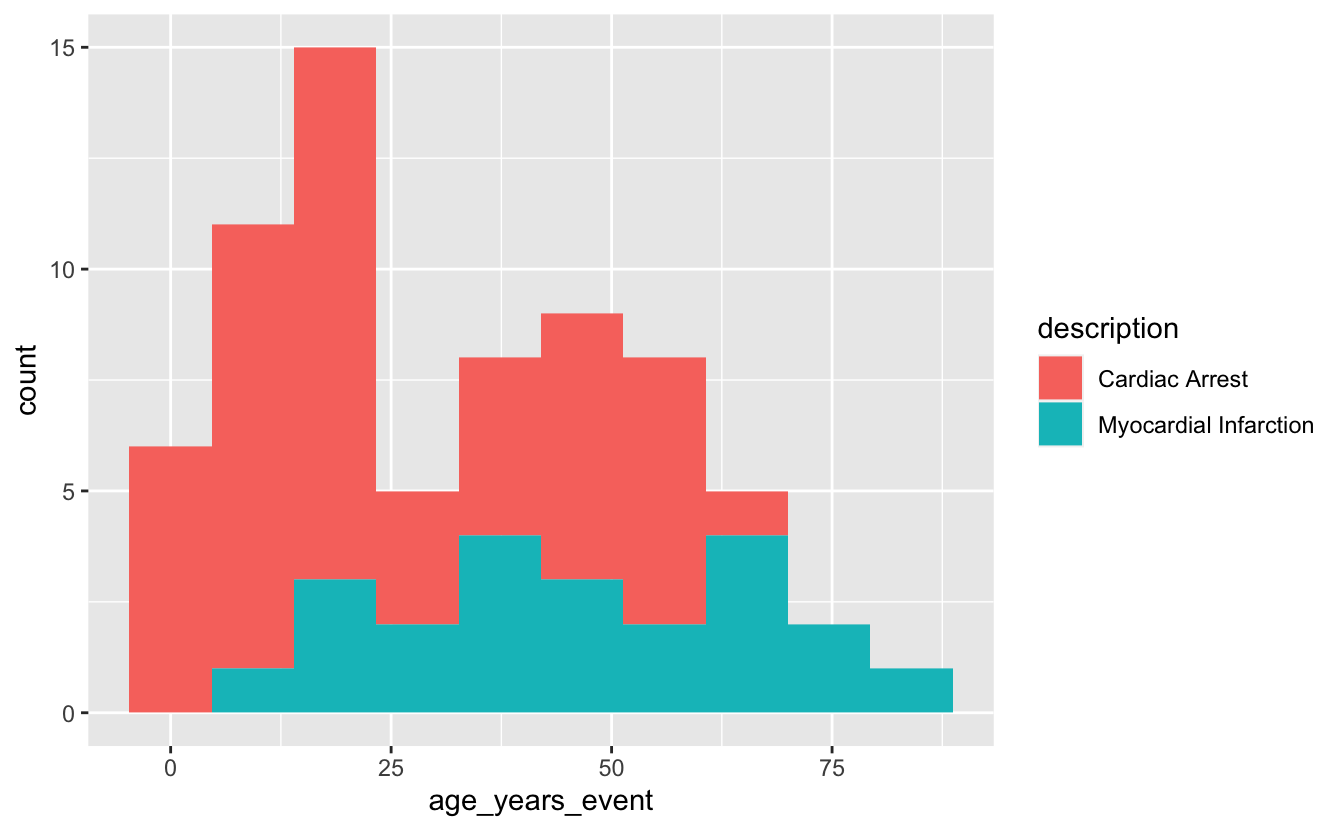
ggplot(mi_encounters_pt, aes(x = age_years_event)) +
geom_histogram(bins = 10) +
facet_wrap(~ description)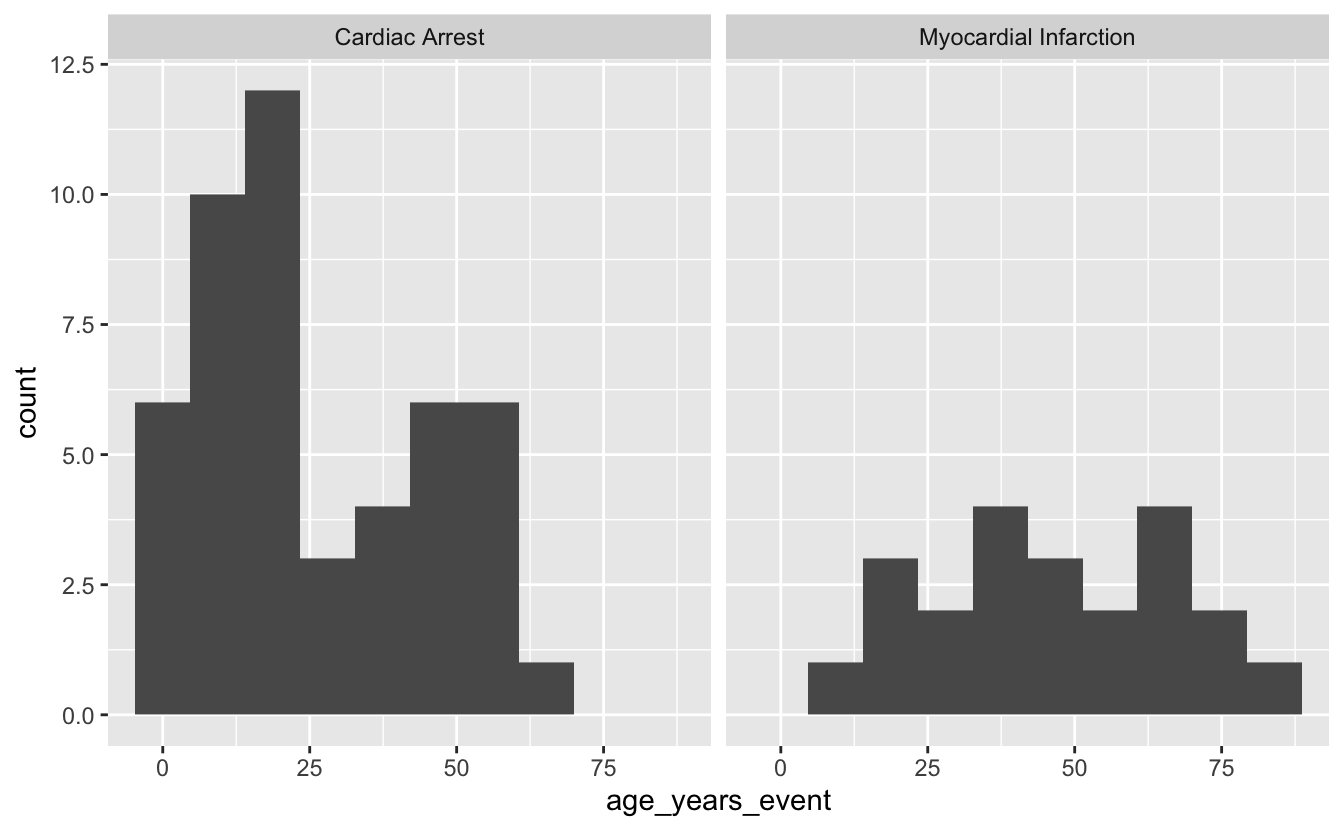
10.2 Databases
https://cran.r-project.org/web/packages/RSQLite/vignettes/RSQLite.html
R
patients <- read_csv("data/synthea/patients.csv")Rows: 1171 Columns: 25── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (19): Id, SSN, DRIVERS, PASSPORT, PREFIX, FIRST, LAST, SUFFIX, MAIDEN, ...
dbl (4): LAT, LON, HEALTHCARE_EXPENSES, HEALTHCARE_COVERAGE
date (2): BIRTHDATE, DEATHDATE
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.encounters <- read_csv("data/synthea/encounters.csv")Rows: 53346 Columns: 15── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (8): Id, PATIENT, ORGANIZATION, PROVIDER, PAYER, ENCOUNTERCLASS, DESCRI...
dbl (5): CODE, BASE_ENCOUNTER_COST, TOTAL_CLAIM_COST, PAYER_COVERAGE, REASO...
dttm (2): START, STOP
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.patients %>%
filter(MARITAL == "M") %>%
select(Id, BIRTHDATE, DEATHDATE, MARITAL) %>%
head(5)# A tibble: 5 × 4
Id BIRTHDATE DEATHDATE MARITAL
<chr> <date> <date> <chr>
1 1d604da9-9a81-4ba9-80c2-de3375d59b40 1989-05-25 NA M
2 034e9e3b-2def-4559-bb2a-7850888ae060 1983-11-14 NA M
3 10339b10-3cd1-4ac3-ac13-ec26728cb592 1992-06-02 NA M
4 8d4c4326-e9de-4f45-9a4c-f8c36bff89ae 1978-05-27 NA M
5 b58731cc-2d8b-4c2d-b327-4cab771af3ef 1970-05-16 NA M 10.2.2 Database tables
R
patients_db <- tbl(con, "patients")
patients_db %>%
filter(MARITAL == "M") %>%
select(Id, BIRTHDATE, DEATHDATE, MARITAL) %>%
head(5)# Source: lazy query [?? x 4]
# Database: sqlite 3.36.0
# [/Users/runner/work/ds4biomed/ds4biomed/data/synthea/synthea.sqlite]
Id BIRTHDATE DEATHDATE MARITAL
<chr> <dbl> <dbl> <chr>
1 1d604da9-9a81-4ba9-80c2-de3375d59b40 7084 NA M
2 034e9e3b-2def-4559-bb2a-7850888ae060 5065 NA M
3 10339b10-3cd1-4ac3-ac13-ec26728cb592 8188 NA M
4 8d4c4326-e9de-4f45-9a4c-f8c36bff89ae 3068 NA M
5 b58731cc-2d8b-4c2d-b327-4cab771af3ef 135 NA M 10.2.3 SQL
R
dbGetQuery(con, 'SELECT * FROM patients LIMIT 5') Id BIRTHDATE DEATHDATE SSN
1 1d604da9-9a81-4ba9-80c2-de3375d59b40 7084 NA 999-76-6866
2 034e9e3b-2def-4559-bb2a-7850888ae060 5065 NA 999-73-5361
3 10339b10-3cd1-4ac3-ac13-ec26728cb592 8188 NA 999-27-3385
4 8d4c4326-e9de-4f45-9a4c-f8c36bff89ae 3068 NA 999-85-4926
5 f5dcd418-09fe-4a2f-baa0-3da800bd8c3a 9787 NA 999-60-7372
DRIVERS PASSPORT PREFIX FIRST LAST SUFFIX
1 S99984236 X19277260X Mr. José Eduardo181 Gómez206 <NA>
2 S99962402 X88275464X Mr. Milo271 Feil794 <NA>
3 S99972682 X73754411X Mr. Jayson808 Fadel536 <NA>
4 S99974448 X40915583X Mrs. Mariana775 Rutherford999 <NA>
5 S99915787 X86772962X Mr. Gregorio366 Auer97 <NA>
MAIDEN MARITAL RACE ETHNICITY GENDER
1 <NA> M white hispanic M
2 <NA> M white nonhispanic M
3 <NA> M white nonhispanic M
4 Williamson769 M white nonhispanic F
5 <NA> <NA> white nonhispanic M
BIRTHPLACE ADDRESS CITY
1 Marigot Saint Andrew Parish DM 427 Balistreri Way Unit 19 Chicopee
2 Danvers Massachusetts US 422 Farrell Path Unit 69 Somerville
3 Springfield Massachusetts US 1056 Harris Lane Suite 70 Chicopee
4 Yarmouth Massachusetts US 999 Kuhn Forge Lowell
5 Patras Achaea GR 1050 Lindgren Extension Apt 38 Boston
STATE COUNTY ZIP LAT LON HEALTHCARE_EXPENSES
1 Massachusetts Hampden County 01013 42.22835 -72.56295 271227.1
2 Massachusetts Middlesex County 02143 42.36070 -71.12653 793946.0
3 Massachusetts Hampden County 01020 42.18164 -72.60884 574111.9
4 Massachusetts Middlesex County 01851 42.63614 -71.34325 935630.3
5 Massachusetts Suffolk County 02135 42.35243 -71.02861 598763.1
HEALTHCARE_COVERAGE
1 1334.88
2 3204.49
3 2606.40
4 8756.19
5 3772.20dbGetQuery(con, 'SELECT Id, BIRTHDATE, DEATHDATE, MARITAL FROM patients WHERE MARITAL = "M" LIMIT 5') Id BIRTHDATE DEATHDATE MARITAL
1 1d604da9-9a81-4ba9-80c2-de3375d59b40 7084 NA M
2 034e9e3b-2def-4559-bb2a-7850888ae060 5065 NA M
3 10339b10-3cd1-4ac3-ac13-ec26728cb592 8188 NA M
4 8d4c4326-e9de-4f45-9a4c-f8c36bff89ae 3068 NA M
5 b58731cc-2d8b-4c2d-b327-4cab771af3ef 135 NA M10.2.3.1 Pass variables to SQL
dbGetQuery(con, 'SELECT Id, BIRTHDATE, DEATHDATE, MARITAL FROM patients WHERE MARITAL = :x LIMIT 5',
params = list(x = "M")) Id BIRTHDATE DEATHDATE MARITAL
1 1d604da9-9a81-4ba9-80c2-de3375d59b40 7084 NA M
2 034e9e3b-2def-4559-bb2a-7850888ae060 5065 NA M
3 10339b10-3cd1-4ac3-ac13-ec26728cb592 8188 NA M
4 8d4c4326-e9de-4f45-9a4c-f8c36bff89ae 3068 NA M
5 b58731cc-2d8b-4c2d-b327-4cab771af3ef 135 NA M10.2.4 Show SQL query
R
patients_db %>%
group_by(MARITAL, RACE, ETHNICITY, GENDER) %>%
summarize(count = n()) %>%
arrange(-count)`summarise()` has grouped output by 'MARITAL', 'RACE', 'ETHNICITY'. You can override using the `.groups` argument.# Source: lazy query [?? x 5]
# Database: sqlite 3.36.0
# [/Users/runner/work/ds4biomed/ds4biomed/data/synthea/synthea.sqlite]
# Groups: MARITAL, RACE, ETHNICITY
# Ordered by: -count
MARITAL RACE ETHNICITY GENDER count
<chr> <chr> <chr> <chr> <int>
1 M white nonhispanic F 259
2 M white nonhispanic M 222
3 <NA> white nonhispanic F 150
4 <NA> white nonhispanic M 135
5 S white nonhispanic F 63
6 S white nonhispanic M 48
7 M black nonhispanic M 31
8 M asian nonhispanic M 23
9 M asian nonhispanic F 22
10 M black nonhispanic F 22
# … with more rowsR
patients_db %>%
group_by(MARITAL, RACE, ETHNICITY, GENDER) %>%
summarize(count = n()) %>%
arrange(-count) %>%
show_query()`summarise()` has grouped output by 'MARITAL', 'RACE', 'ETHNICITY'. You can override using the `.groups` argument.<SQL>
SELECT `MARITAL`, `RACE`, `ETHNICITY`, `GENDER`, COUNT(*) AS `count`
FROM `patients`
GROUP BY `MARITAL`, `RACE`, `ETHNICITY`, `GENDER`
ORDER BY -`count`R
dbGetQuery(con,
"SELECT `MARITAL`, `RACE`, `ETHNICITY`, `GENDER`, COUNT(*) AS `count`
FROM `patients`
GROUP BY `MARITAL`, `RACE`, `ETHNICITY`, `GENDER`
ORDER BY -`count`") MARITAL RACE ETHNICITY GENDER count
1 M white nonhispanic F 259
2 M white nonhispanic M 222
3 <NA> white nonhispanic F 150
4 <NA> white nonhispanic M 135
5 S white nonhispanic F 63
6 S white nonhispanic M 48
7 M black nonhispanic M 31
8 M asian nonhispanic M 23
9 M asian nonhispanic F 22
10 M black nonhispanic F 22
11 <NA> white hispanic M 21
12 M white hispanic M 20
13 <NA> white hispanic F 18
14 M white hispanic F 15
15 <NA> black nonhispanic M 14
16 <NA> black nonhispanic F 12
17 <NA> asian nonhispanic F 11
18 <NA> asian nonhispanic M 10
19 M native nonhispanic M 9
20 S white hispanic M 9
21 S asian nonhispanic M 8
22 S black nonhispanic M 7
23 M asian hispanic F 5
24 M black hispanic F 5
25 S black nonhispanic F 5
26 S white hispanic F 5
27 S asian nonhispanic F 4
28 <NA> black hispanic F 3
29 <NA> asian hispanic F 2
30 <NA> asian hispanic M 2
31 M native hispanic F 2
32 S asian hispanic F 2
33 <NA> native nonhispanic M 1
34 <NA> other nonhispanic F 1
35 M asian hispanic M 1
36 M black hispanic M 1
37 M other hispanic F 1
38 S black hispanic F 1
39 S native nonhispanic F 1